|
Selected/Multiple reaction monitoring assays, conducted on triple quadrupole instruments, can be coupled to liquid chromatography for the analysis of complex proteome digests.
In SRM/MRM assays the first (Q1) and last (Q3) mass analyzers of a triple quadrupole mass spectrometer are used as mass filters to isolate a peptide ion and a corresponding fragment ion. The signal of the fragment ion is then monitored over the chromatographic elution time (Fig. 1). The selectivity resulting from the two filtering stages, combined with the high duty cycle, results in quantitative analyses with unmatched sensitivity and specificity. The specific pairs of m/z values associated to the precursor and fragment ions selected are referred to as "transitions" and effectively constitute mass spectrometric assays that allow to identify and quantify a specific peptide and, by inference, the corresponding protein in a complex protein digest.
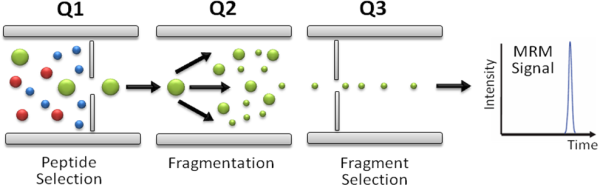
Figure 1.
The process of establishing a SRM/MRM assay for a protein consists of a number of steps:
1. Selection of the appropriate peptide/s, unique to the protein of interest and showing high mass spectrometry signal response (proteotypic peptides, PTPs), to maximize the sensitivity of the assay
2. Selection of predominant peptide fragments specific to the peptide of interest to be used in the MRM transition
3. Eventually, for each peptide-fragment pair, optimization of specific MS parameters (e.g. the collision energy) to maximize the signal response/sensitivity
4. Validation of the MRM assay to confirm peptide identity, e.g. by acquiring a full MS2 spectrum of the peptide in the triple quadrupole instrument used for MRM
5. Extraction of the final “coordinates” of the MRM assay, including the selected peptide and peptide fragments, the corresponding mass-to-charge ratios, the fragment intensity ratios, the associated collision energy, and the chromatographic elution time to be optionally used in time-constrained MRM analyses
Overall, this is a lengthy and iterative process, but, once an MRM assay for a protein is established, it becomes universally useful, i.e. the tedious assay development process needs to be performed only once, for a given type of mass spectrometer and fragmentation mechanism (e.g. collision induced-dissociation).
In the MRMAtlas each protein assay is presented as a set of optimal MRM coordinates for the peptide(s) that represent a protein. Peptide identifications have been validated by acquiring the corresponding tandem mass spectra on the triple quadrupole mass spectrometer, which can be viewed as single or consensus spectra. The final assay coordinates can be directly downloaded in Excel table-format which can be directly pasted into a MRM/SRM method of a triple quadrupole instrument and used to specifically detect and quantify the protein of interest in a complex protein digest.
(Adapted from: A. Schimdt, P. Picotti and R. Aebersold Proteomeanalyse und systembiologie BIOspektrum, 1/2008, S. 44)
|
